DNA Assembly
with Synthace
Transform combinatorial assembly designs into flexible automation instructions and dynamic liquid calculations—all without writing any code.
Request a Demo Take a tour
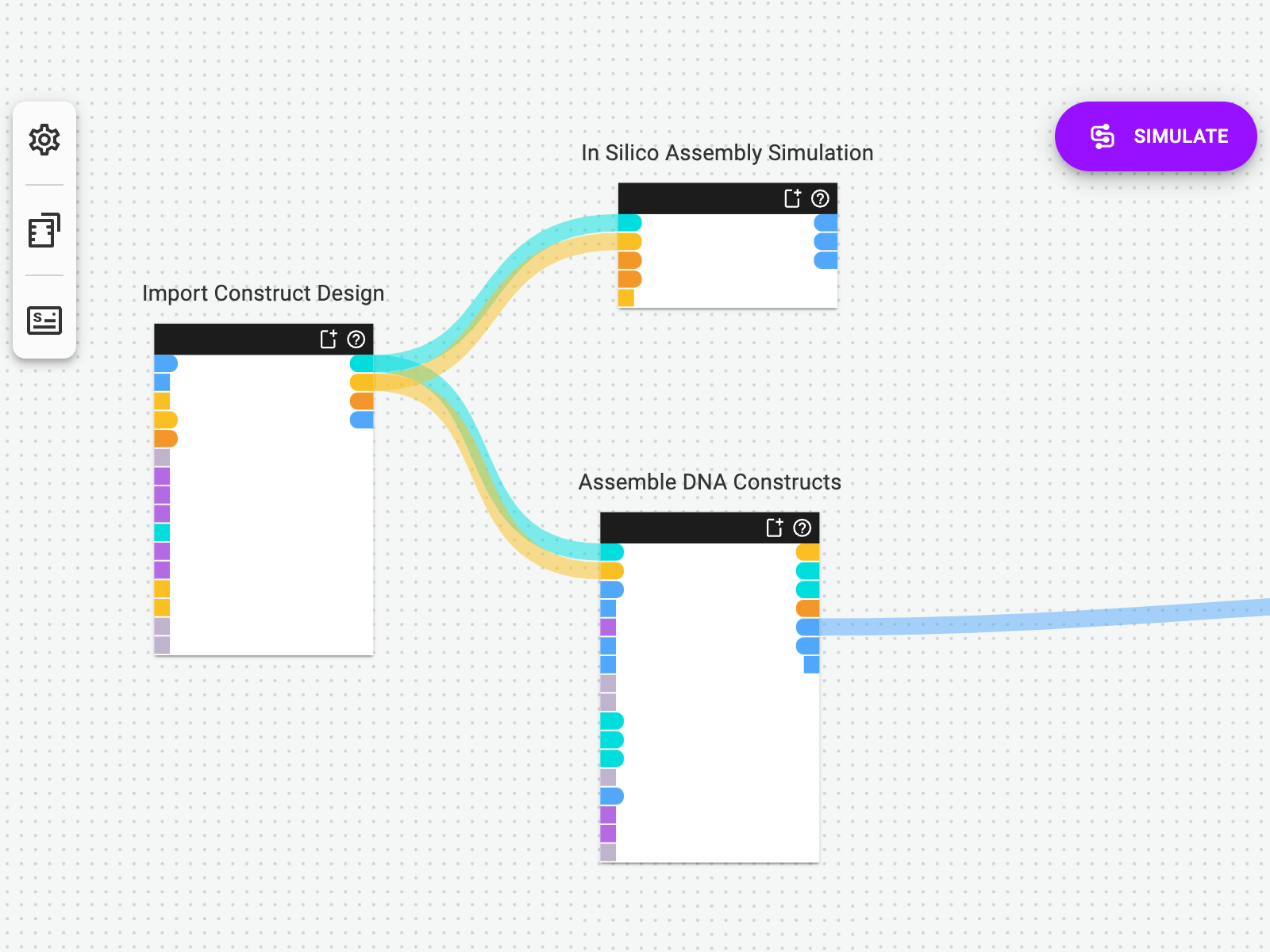
Import a Design File to Get Started
Synthace lets you import your assembly design files into the platform at the beginning of your workflow. Once you’ve specified the plasmids you want to make, the platform will perform all of the combinatorial calculations and instructions you needed to automate your cloning.
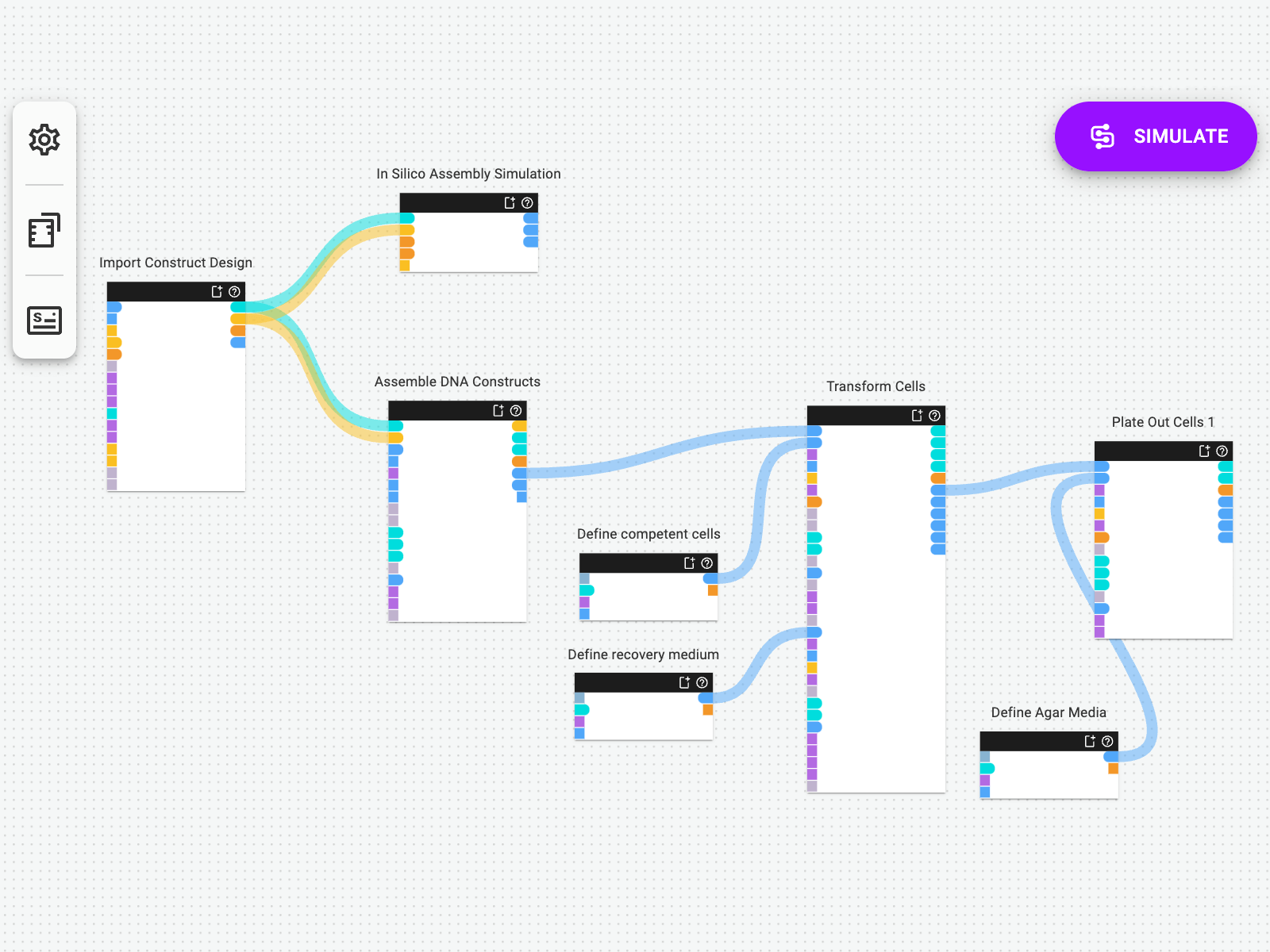
Re-Use Protocols Again and Again
Design one protocol once, and simply upload new design files for each set of DNA assemblies you want to execute. With Synthace’s dynamic liquid handler programming, there’s no need to reinvent the wheel: new designs are re-calculated and automated as often as you need them.
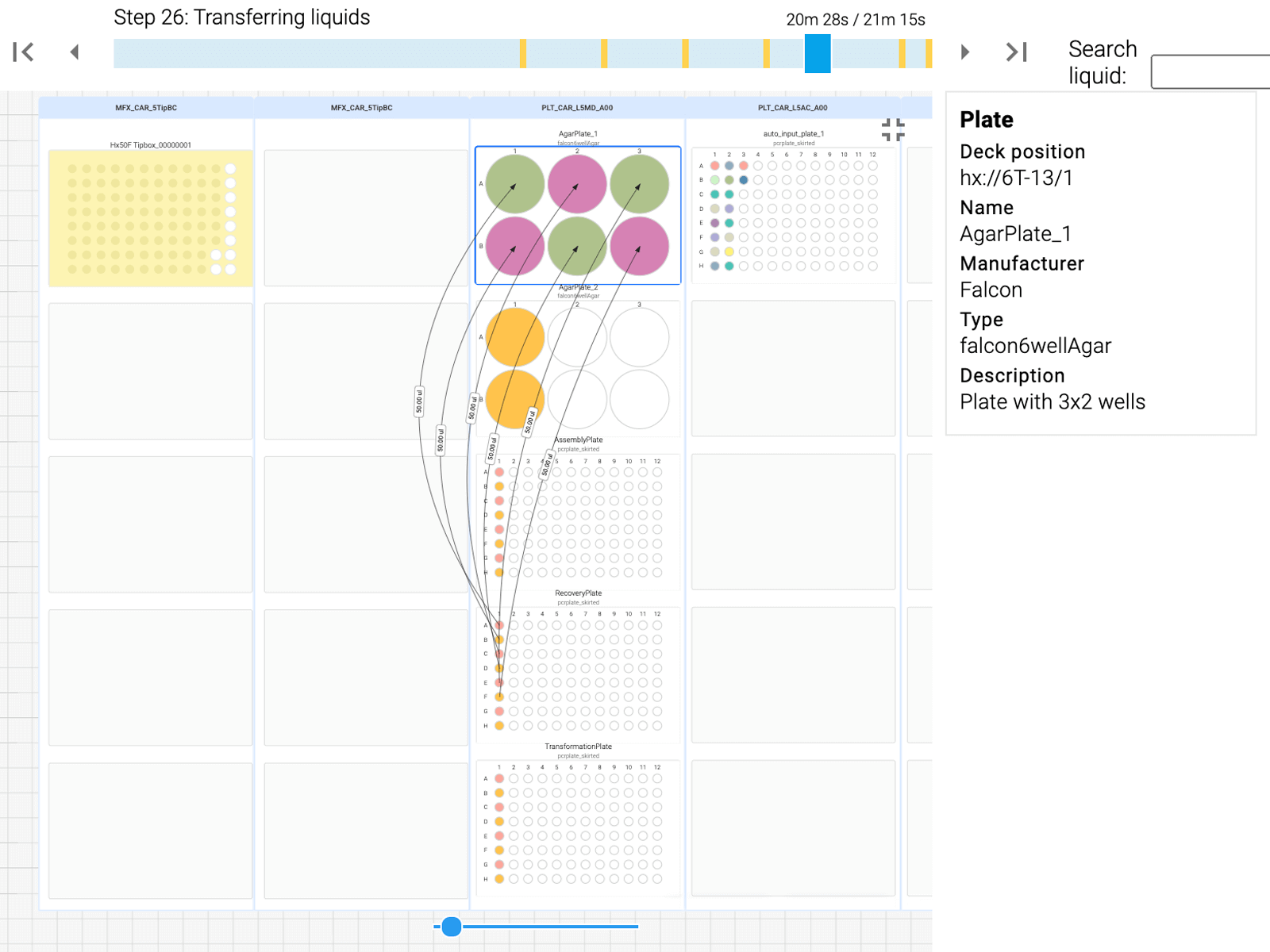
Advanced Calculations done for you In Silico
Specify the plasmids you’re aiming for and the fragments you’ll be using and Synthace will calculate all of the liquid movements, concentrations and dilutions needed throughout the entire process. It then translates this into automation instructions and runs this on your equipment.
The Latest From Our Blog
View All PostsMeet Professor Chris Molloy, CEO at Medicines Discovery Catapult

Meet Thierry Dorval, Head of Data Science and Management at Servier

Meet Dan Thomas, CEO & Founding Consultant at Twenty24 Consulting







